Fragment Analysis Service
The GUDSF provides a fragment analysis service using the Applied Biosystems SeqStudio 8 Flex. The facility’s SeqStudio 8 Flex has a 50cm array and POP-7 polymer installed
Applications include:
- MLPA
- Microsatellite analysis
- AFLP
- SNaPshot® SNP analysis
- Cell Line Authentication
- Custom analyses
The fragment analysis service provides capillary separation and size determination of fluorescent-labelled DNA fragments. There are two service options available. For each option, the customer performs the fluorescent labelling PCR and prepares the reaction products for electrophoresis.
To assess the accuracy of the electrophoretic analysis and subsequent DNA fragment sizing analysis, an internal control sample is run with each batch. The internal control samples are the GeneScan DS-33 Installation Standard or GeneScan DS-30 Installation Standard.
Service Options
- Ready-to-Load Samples
This service requires the customer to perform the PCR reaction and submit the pre-mixed labelled DNA (reaction products) in Size Standard/HiDi Formamide ready for capillary separation & analysis.
- Amplification products
This service requires the customer to perform the PCR reaction and submit an aliquot of diluted, labelled DNA reaction products. The GUDSF will add the size standard & HiDi Formamide.
The GUDSF accepts samples labelled with dyes belonging to the following dye sets. Multiplexing is accommodated using dyes within each dye set. The GUDSF only stocks the LIZ600v2 size standards. For the DS30 & Promega kits, the customer must supply the size standard. Contact the GUDSF for further information.
- DS33/G5: 6-FAM, VIC, NED, PET and (LIZ reserved for the size standard)
- DS30/D: 6-FAM, HEX, NED and (ROX reserved for our size standard)
- DS36/J6: 6-FAM, VIC, NED, SID, TAZ and (LIZ reserved for the size standard)
- DS37/J6-T: 6-FAM, VIC, TED, SID, TAZ and (LIZ reserved for the size standard).
- Promega Powerplex® Kits using the 4C, 5C or 6C matrix standard.
Sample Submission
- An iLab Request must accompany each sample submission.
- Submit at least 1µL of diluted product in a 02.mL strip-tube or 96-well plates.
- For Ready-to-Load samples, please supply samples (1µL sample + 9µL size standard/HiDi mix) in a 96-well plate (MicroAmp Optical Reaction or Axygen half skirt plate).
- It is recommended that samples be shipped on ice (4oC).
- Wrap sample batches in alfoil for protection (light sensitive).
- For samples submitted in full, 96-well plates please leave well position A01 empty for the internal control sample.
- Clearly mark the orientation of the plate e.g. Top left corner = well A01 and label the plate.
- Simplify your labels, for example GUS001, GUS002 etc.
View your data
A data file is created for each sample (.fsa). To analyse and view the data GeneMapper or Peak Scanner (free) software is available from Life Technologies.
Options for analysis and results delivery include allele calling, tabulated data or PDF prints.
Customer Support
For customer support and troubleshooting advice please contact GUDSF staff.
For fragment analysis application and experimental design information, please refer to the DNA Fragment Analysis User Guide
Data Archive
The GUDSF maintains an archive of customer data, however customers are advised to back-up their data. If you require a copy of a past result please contact the GUDSF.
Experimental design
A good source of information on experimental design, primer labelling, optimising PCR, multiplexing reactions, fragment analysis applications and troubleshooting, is the DNA Fragment Analysis by Capillary Electrophoresis, Applied Biosystems User Guide.
Multiplexing Reactions
Using the SeqStudio 8 Flex and the Applied Biosystems GeneMapper system, you can label different DNA fragments with up to five different coloured fluorescent dyes depending upon the selected dye set. To exploit the potential for increased throughput using this system, you may want to multiplex electrophoresis by co-loading products of multiple PCR reactions in the same capillary injection or depending upon your application, you may choose to multiplex the PCR.
Co-electrophoresis
- Use a combination of dyes from Applied Biosystems Dye Set G5, DS31 or DS30. (LIZ or ROX is reserved for our internal standard).
- Use different dye labels for PCR reactions with overlapping product size.
- Pool PCR products in order to get similar fluorescent intensities for all products. The intensity of emitted fluorescence is different for each dye, 6-FAM, VIC >NED > PET > LIZ, so use a greater amount of PCR product labelled with dyes of low emission intensity than those labelled with dyes of high emission intensity.
Multiplexing PCR
- Combine more than one pair of primers in the same PCR reaction tube.
- Do not multiplex primers with similar product lengths labelled with similar dyes.
- For microsatellite applications, do not multiplex same dye-labelled primers for loci with overlapping allele size ranges.
- Check for compatibility between the primers.
- Test for successful co-amplification between primer pairs.
Sizing
When designing your primers ensure that the fragments produced are >75 bp and < 490bp (for LIZ500) or < 580bp (for LIZ600). It is critical to the sizing method employed by the GeneMapper software that there are at least two size standard fragments larger than your largest unknown fragment.
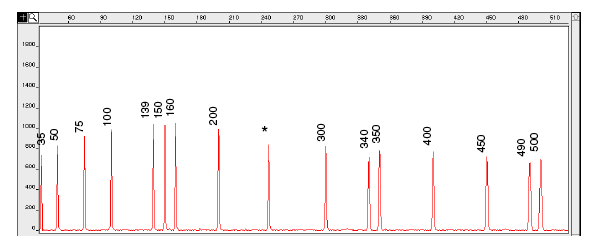
Figure 5-5: Electropherogram of the GeneScan-500 size standard run under denaturing conditions on the ABI PRISM 310 Genetic Analyser. Fragments were run using the POP-4 polymer at 60°C.
IMPORTANT An * for the 250-bp peak denotes a peak resulting from abnormal migration of double strands that did not completely separate under denaturing conditions. Do not use this peak to size samples. This peak shows variably smaller values than the actual size of the fragments.
Control DNA
It is recommended that you analyse at least one control DNA sample in every PCR run.
Control DNA :
- Serves as a positive control for troubleshooting problems with the PCR amplification
- Allows you to monitor sizing precision
- Enables you to correlate the fragment sizes that you obtain with the fragment sizes obtained by others.
Sample Preparation
It is the responsibility of the customer to optimise the concentration of reaction products in the sample. You will almost always need to dilute PCR amplification products before sending to the GUDSF for analysis. Typically, the required dilution lies in the range from 1:3 – 1:80 ( PCR product:distilled H2O).
Too much signal is the most common problem. For optimal results, the fluorescent signal should be between 150 – 6000 RFUs . Above this range, the instrument cannot measure the true value of the signal and therefore cannot apply the matrix correctly. This results in artefact “pull-up” peaks that can appear in other colours. Artefact peaks can corrupt both the automated size calling and the analysis of co-loaded samples.
If you intend to pool PCR products, it is important to pool PCR products together at the correct ratios in order to get similar fluorescent intensities across all fragments in the pool. The fluorescent dyes are detected with different efficiencies, therefore the amount of each dye-labelled product in the pool will require adjustment to ensure even detection.
A good way to proceed is to test a few combinations of pooled PCR reactions to determine the pooling ratio that will provide similar fluorescent intensities across all the pooled fragments. Then carry out a series of dilutions on the pooled reactions in order to determine the optimal fluorescence for running on the 3130xl.
Send in 1 µL of each dilution series to the GUDSF for capillary electrophoresis and sizing. After determining the optimal pooling ratio and/or dilution ratio, you can then use the same dilutions for subsequent analyses, as PCR yields should be relatively consistent.
Re-run Policy
Off-scale samples will not be repeated as a rule. The customer is advised to dilute the sample. Customers are further recommended to trial dilutions at the start of each project. Where a customer has requested a sample dilution and re-run, a charge will apply.
Occasionally, a sample may exhibit early loss of resolution or some other capillary related problem. In these instances, the sample is re-run without charge to the customer.